mosquito单细胞测序应用:Nature 人类肝细胞图谱
2021-05-19 来源:腾泉生物 点击次数:4325
德国马克斯普朗克研究所的Dominic Grün团队与法国斯特拉斯堡大学的Thomas F. Baumert团队成功地构建出完整的人类肝脏细胞精细图谱,该研究成果发表于《Nature》杂志上,论文题目为A human liver cell atlas reveals heterogeneity and epithelial progenitors。
研究方法:
该团队使用mCEL-Seq2方法对来自9个健康人类肝脏组织的10000多个细胞进行了深度单细胞测序分析,绘制的图谱涵盖了所有重要的肝脏细胞类型,包括肝实质细胞、血管内皮细胞、巨噬细胞以及其他免疫细胞类型,并且找到了多种新型肝脏细胞。
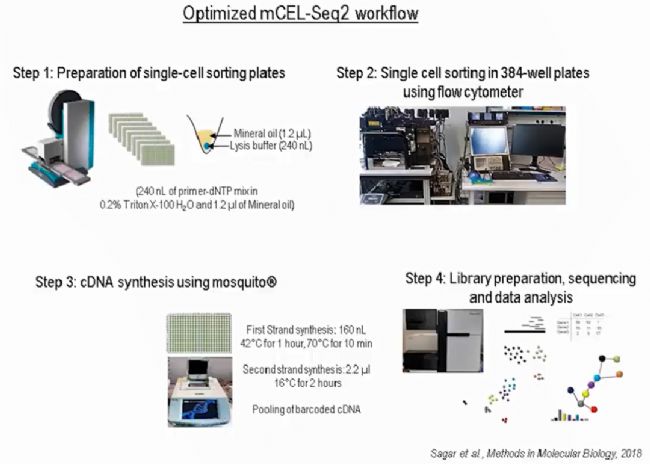
肝细胞悬液通过流式系统把单个细胞分选到384孔板,基于微孔板的mCEL-Seq2单细胞测序步骤由mosquito纳升级单细胞建库系统完成。
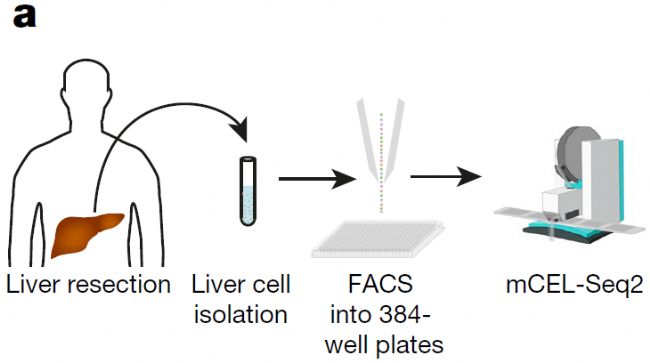
mCEL-Seq2方法在cDNA第一链合成的过程中给每个细胞都加入了barcoded primer,通过体外转录(IVT)方式扩增单细胞cDNA,这既保证了检测方法的灵敏度,又通过把所有样本pooling后再做建库的方式降低了检测成本。通过mosquito纳升级单细胞建库系统的加持,再把反应体系缩小了5倍,把成本压到最低!
Materials&Methods
- mosquito LV, 384-well plates
- cells sorted into 240 nL lysis buffer
- 160 nL RT mix added
- cDNA synthesis in 400 nL total
- using Vapor-Lock to block evaporation
- add cell barcodes; pooled library prep
研究结果:
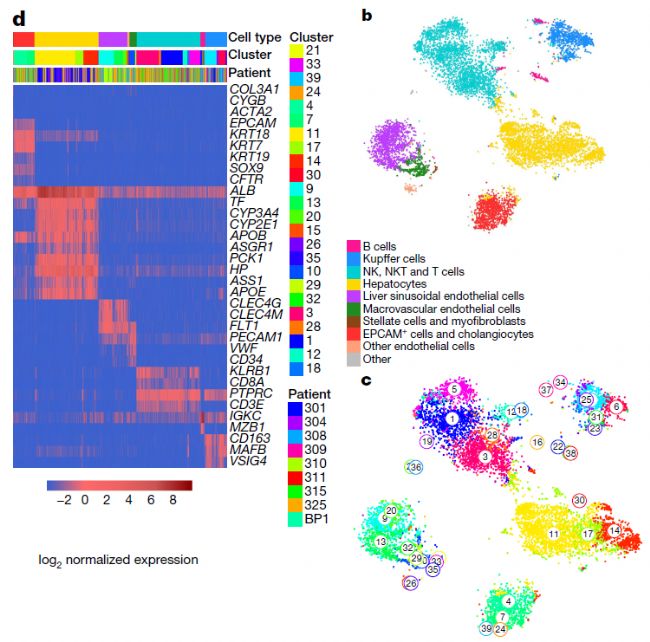
该图谱根据marker基因的表达水平鉴定了几乎所有的肝脏细胞类型,包括肝细胞、EPCAM+胆管细胞(胆管细胞)、CLEC4G+肝血源性内皮细胞(LSECs)、CD34+ PECAM high大血管内皮细胞(MaVECs)、肝星状细胞和肌成纤维细胞、Kupffer细胞和免疫细胞。
研究人员同时发现了从未被报道过的新的肝脏细胞类型。这些细胞亚型虽然在形态上与普通的肝脏细胞没有什么不同,但在基因表达上却截然不同。这些发现归功于单细胞测序实验方法mCEL-seq2和分析算法FateID的重大进展,通过这些方法得以对单个细胞进行高分辨率的深度分析。
参考文献:
Aizarani N , Saviano A , Sagar, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors[J]. Nature, 2019, 572(7768):199-204.
Sagar, Herman J S , Pospisilik J A , et al. High-Throughput Single-Cell RNA Sequencing and Data Analysis[M]. 2018.
Herman J S , Sagar, D Grün. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data[J]. Nature Methods, 2018.
DNA Damage Signaling Instructs Polyploid Macrophage Fate in Granulomas.[J]. Cell, 2018.
德国马普所的自动化CEL-Seq2流程,节省5倍成本

相关文章
更多 >

